- 09 Oct 2025
- 5 Minutes to read
- Print
- DarkLight
Plaso
- Updated on 09 Oct 2025
- 5 Minutes to read
- Print
- DarkLight
Plaso Extension Pricing
While it is free to enable the Plaso extension, pricing is applied to both the original downloaded artifact and the processed (Plaso) artifacts -- $0.02/GB for the original downloaded artifact, and $1.0/GB for the generation of the processed artifacts.
About
Plaso is a Python-based suite of tools used for creation of analysis timelines from forensic artifacts acquired from an endpoint.
These timelines are invaluable tools for digital forensic investigators and analysts, enabling them to effectively correlate the vast quantities of information encountered in logs and various forensic artifacts encountered in an intrusion investigation.
The primary tools in the Plaso suite used for this process are log2timeline, psort, and psteal.
log2timeline- bulk forensic artifact parserpsort- builds timelines based on output fromlog2timelinepsteal- Simply a wrapper forlog2timelineandpsort
The ext-plaso extension within LimaCharlie allows you to run log2timeline and psort (using the psteal wrapper) against artifacts obtained from an endpoint, such as event logs, registry hives, and various other forensic artifacts. When executed, Plaso will parse and extract information from all acquired evidence artifacts that it has support for. Supported parsers are found here.
Extension Configuration
Long Execution Times
Note that it can take several minutes for the plaso generation to complete for larger triage collections, but once it finishes you will see the results in the ext-plaso Sensor timeline, as well as the uploaded artifacts on the Artifacts page.
The ext-plaso extension runs psteal (log2timeline + psort) against the acquired evidence using the following commands:
psteal.py --source /path/to/artifact -o dynamic --storage-file $artifact_id.plaso -w $artifact_id.csv
Upon running psteal.py, a .plaso file and a .csv file are generated. They will be uploaded as LimaCharlie artifacts.
Resulting
.plasofile contains the raw output oflog2timeline.pyResulting
.csvfile contains the CSV formatted version of the.plasofile contents
pinfo.py $artifact_id.plaso -w $artifact_id_pinfo.json --output_format json
After psteal.py runs, information is gathered from the resulting .plaso file using the pinfo.py utility and pushed into the ext-plaso sensor timeline as a pinfo event. This event provides a detailed summary with metrics of the processing that occurred, as well as any relevant errors you should be aware of.
The following events will be pushed to the ext-plaso sensor timeline:
job_queued: indicates thatext-plasohas received and queued a request to process datajob_started: indicates thatext-plasohas started processing the datapinfo: contains thepinfo.pyoutput summarizing the results of the plaso file generationplaso: contains theartifact_idof the plaso file that was uploaded to LimaCharliecsv: contains theartifact_idof the CSV file that was uploaded to LimaCharlie
Usage & Automation
LimaCharlie can automatically kick off evidence processing with Plaso based off of the artifact ID provided in a rule action, or you can run it manually via the extension.
Velociraptor Triage Acquisition Processing
If you use the LimaCharlie Velociraptor extension, a good use case of ext-plaso would be to trigger Plaso evidence processing upon ingestion of a Velociraptor KAPE files artifact collection.
Configure a D&R rule to watch for Velociraptor collection events upon ingestion, and then trigger the Plaso extension:
Detect:
op: and target: artifact_event rules: - op: is path: routing/log_type value: velociraptor - op: is not: true path: routing/event_type value: export_completeRespond:
- action: extension request extension action: generate extension name: ext-plaso extension request: artifact_id: '{{ .routing.log_id }}'Launch a
Windows.KapeFiles.Targetsartifact collection in the LimaCharlie Velociraptor extension. This instructs Velociraptor to gather all endpoint artifacts defined in this KAPE Target file.Argument options:
EventLogs=Y- EventLogs only, quicker processing time for proof of conceptKapeTriage=Y- full KapeTriage files collection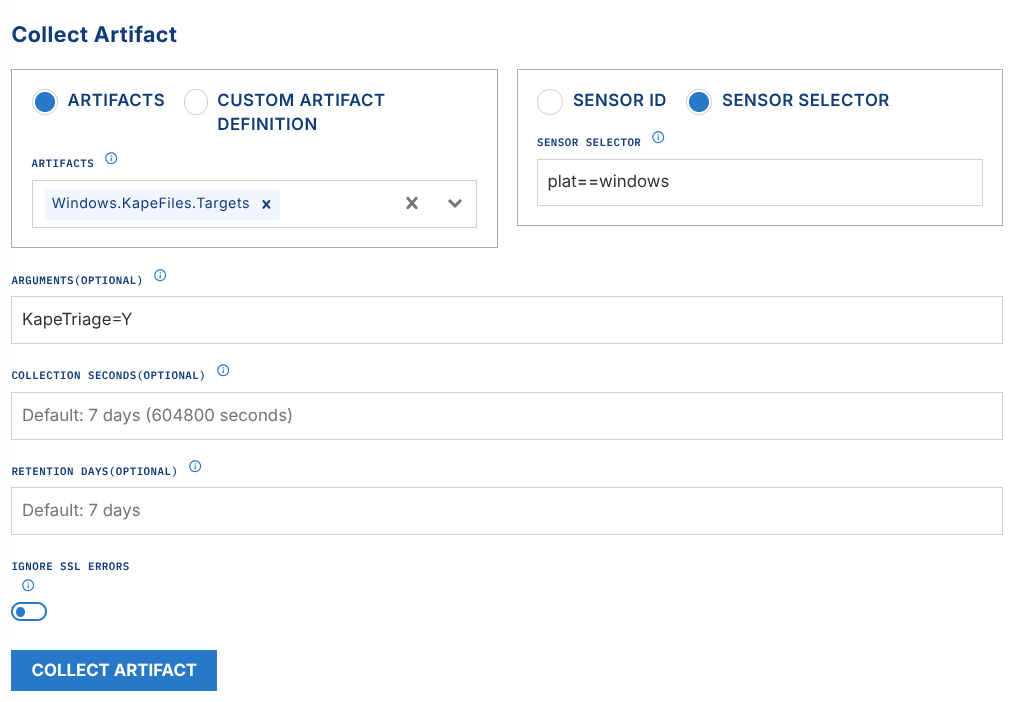
Once Velociraptor collects, zips, and uploads the evidence, the previously created D&R rule will send the triage
.ziptoext-plasofor processing. Watch theext-plasosensor timeline for status and the Artifacts page for the resulting.plaso&.csvoutput files. See Working with the Output.
MFT Processing
If you use the LimaCharlie Dumper extension, a good use case of ext-plaso would be to trigger Plaso evidence processing upon ingestion of a MFT CSV artifact.
Configure a D&R rule to watch for MFT collection events upon ingestion, and then trigger the Plaso extension:
Detect:
op: and target: artifact_event rules: - op: is path: routing/log_type value: mftcsv - op: is not: true path: routing/event_type value: export_completeRespond:
- action: extension request extension action: generate extension name: ext-plaso extension request: artifact_id: '{{ .routing.log_id }}'Launch an MFT dump in the LimaCharlie Dumper extension.
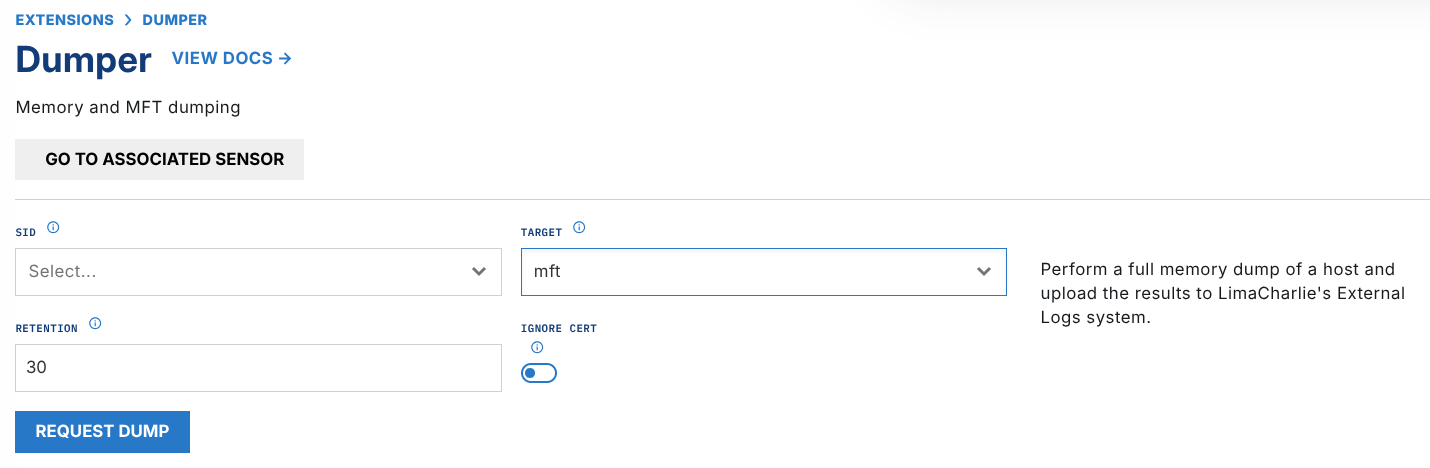
Once dumper is complete and uploads the evidence, the previously created D&R rule will send the zipped MFT CSV to
ext-plasofor processing. Watch theext-plasosensor timeline for status and the Artifacts page for the resulting.plaso&.csvoutput files. See Working with the Output.
Working with the Output
Running the extension generates the following useful outputs:
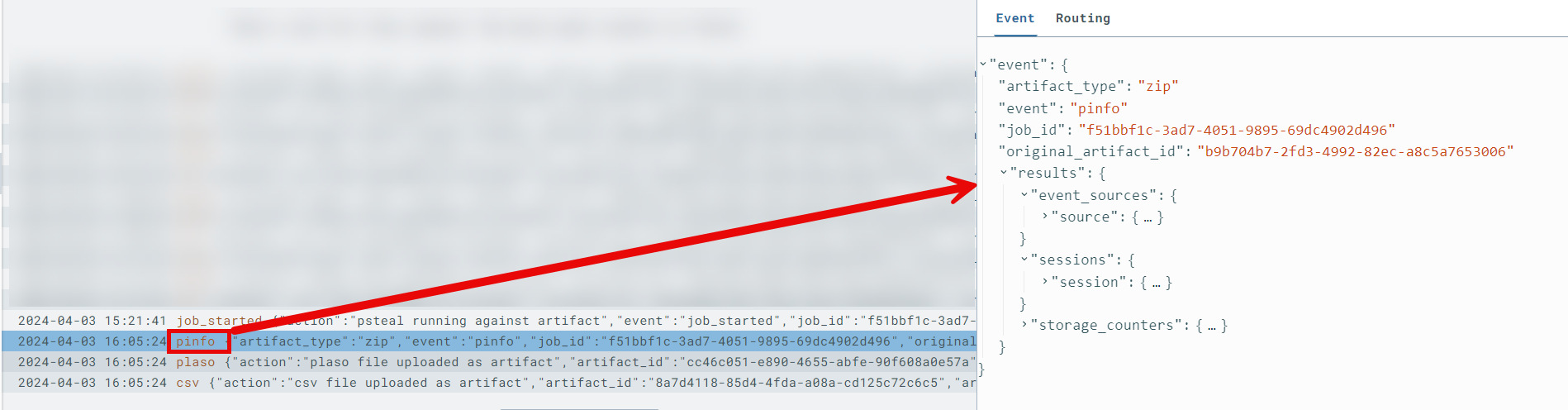
pinfoonext-plasosensor timeline
First and foremost, after the completion of a processing job byext-plaso, it is highly encouraged to analyze the resultingpinfoevent on theext-plasosensor timeline. This event provides a detailed summary with metrics of the processing that occurred, as well as any relevant errors you should be aware of.Pay close attention to fields such as
warnings_by_parserorwarnings_by_path_specwhich may reveal parser errors that were encountered.Sample output of
pinfoshowing counts of parsed artifacts nested understorage_counters-- this provides insight as to which, and how many events will be present in your CSV timeline.
"amcache": 986,
"appcompatcache": 4096,
"bagmru": 29,
"chrome_27_history": 29,
"chrome_66_cookies": 246,
"explorer_mountpoints2": 2,
"explorer_programscache": 1,
"filestat": 3495,
"lnk": 160,
"mft": 4790977,
"mrulist_string": 2,
"mrulistex_shell_item_list": 3,
"mrulistex_string": 5,
"mrulistex_string_and_shell_item": 5,
"mrulistex_string_and_shell_item_list": 1,
"msie_webcache": 143,
"msie_zone": 60,
"networks": 4,
"olecf_automatic_destinations": 37,
"olecf_default": 5,
"recycle_bin": 3,
"shell_items": 297,
"total": 5840430,
"user_access_logging": 34,
"userassist": 44,
"utmp": 13,
"windows_boot_execute": 8,
"windows_run": 10,
"windows_sam_users": 16,
"windows_services": 2004,
"windows_shutdown": 8,
"windows_task_cache": 835,
"windows_timezone": 4,
"windows_typed_urls": 3,
"windows_version": 6,
"winevtx": 382674,
"winlogon": 8,
"winreg_default": 654177
Downloadable Artifacts

plasoartifact
The downloadable.plasofile contains the raw output oflog2timeline.pyand can be imported into Timesketch as a timeline.csvartifact
The downloadable.csvfile can be easily viewed in any CSV viewer, but a highly recommended tool for this is Timeline Explorer from Eric Zimmerman.

